Page History
...
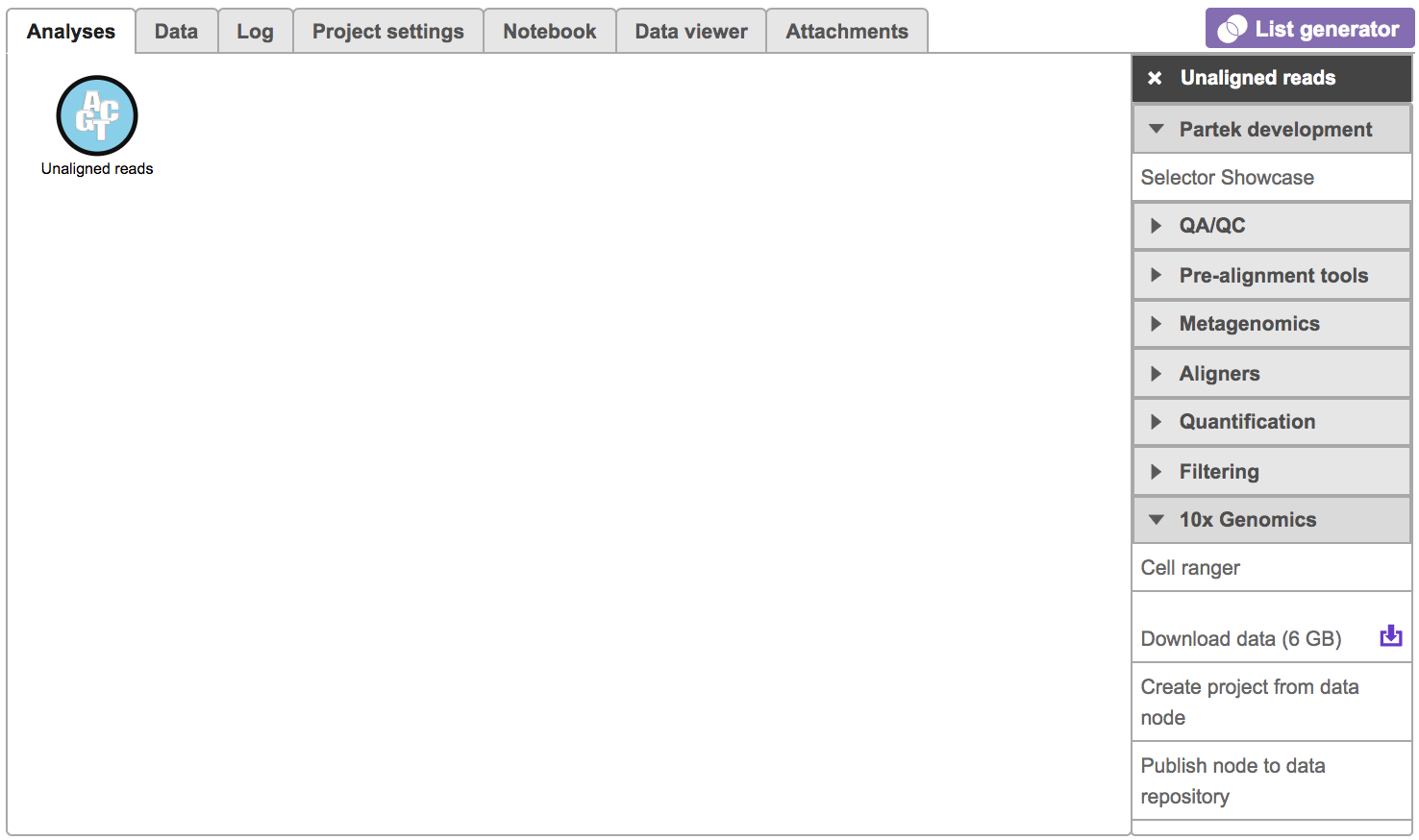
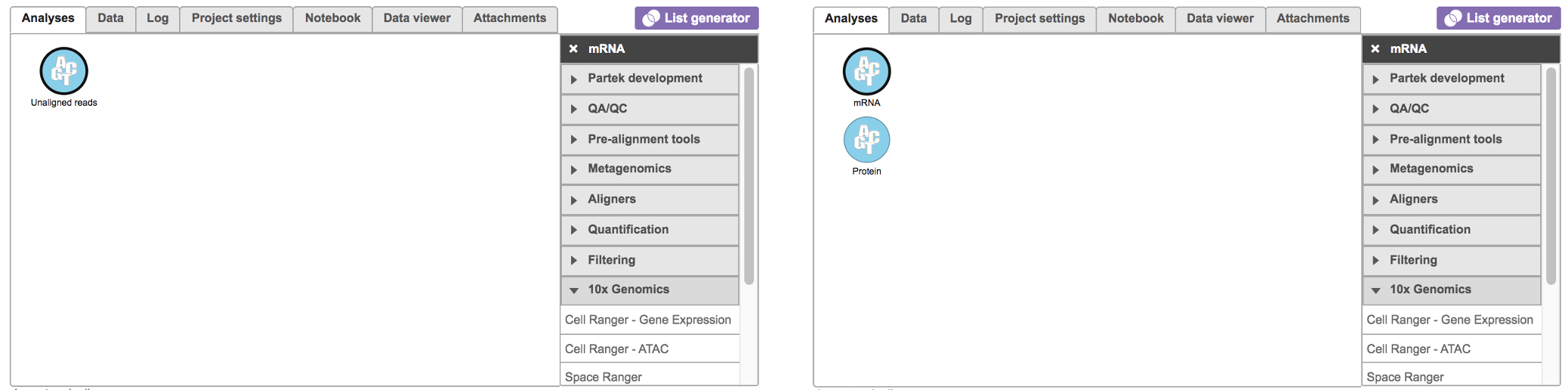
To run the Cell Ranger - Gene Expression task for scRNA-seq data in Flow, select Unaligned reads datanode, then select Cell Ranger - Gene Expression in the 10x Genomics section (left panel, Figure 1). For Feature Barcode data, there will be two datanodes once the fastq files imported in Flow properly - mRNA and protein (right panel, Figure 1). Users should select mRNA datanode to trigger the Cell ranger - Gene Expression task.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
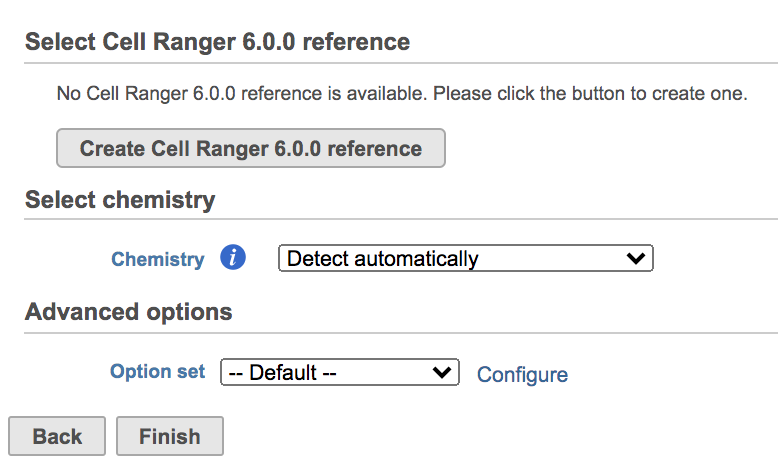
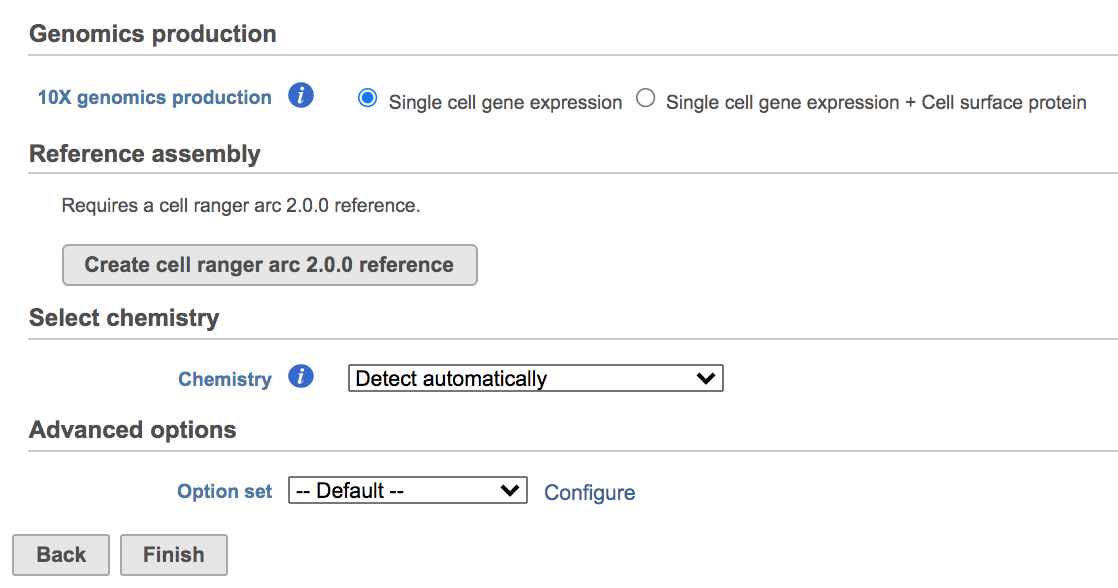
Once the Genomics production has been picked, users will be asked to create a Cell Ranger 6.0.0 reference genome Reference assembly if it is the first time to run the Cell Ranger - Gene Expression task in Flow (Figure 2). In Partek Flow, we will use Cell Ranger ARC 2.0.0 to create reference assembly for all 10x Genomics analysis pipelines. To create and use a reference assembly, Cell Ranger ARC requires a reference genome sequence (FASTA file) and gene annotations (GTF file).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Clicking the big grey button of Create Cell Ranger 6.0.0 reference would pop up a new window where lists the three pre-built reference genomes for human(hg38), mouse(mm10) and the mix of two(hg38-mm10), respectively (Figure 3). They are exactly the same reference genomes (2020-A) that are provided in Cell Ranger by default. In details, the transcriptome annotations are respectively GENCODE v32 for human and vM23 for mouse, which are equivalent to Ensembl 98[3]. References for other organisms currently are not available in Flow Cell ranger, and will be coming in the future.
...