Page History
...
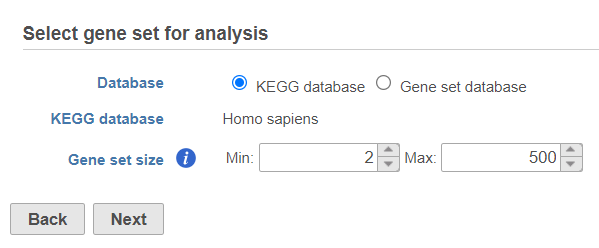
Use the first dialog (Figure 2) to specify the gene sets. You can run GSEA on pathways (currently based on Kyoto Encyclopedia of Genes and Genomse (KEGG) pathways) or on other gene set databases. When using the KEGG option, the KEGG database is automatically set, based on the upstream nodes. The Gene set size options allows you to restrict your analysis on gene sets of certain size (i.e. number of genes).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
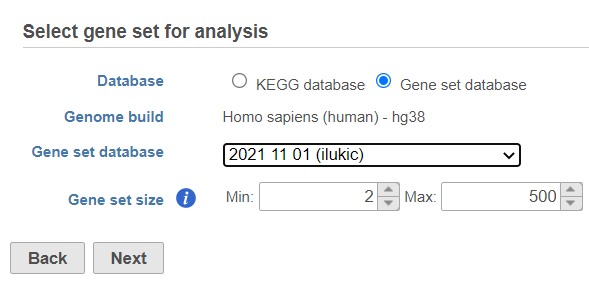
If you select Gene set database option, another , two additional options will appear. Genome build will be detected automatically, based on the upstream nodes. The gene sets that are available for that build are listed in the drop down list (Figure 3). Custom databases will be labeled by their name as specified in the Library file management, while GO database will be labeled by the release date (as seen in Figure 3).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
oiioioio
Once your choices are made, push Next to proceed.
References
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545-15550. doi:10.1073/pnas.0506580102
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267-273. doi:10.1038/ng1180