Page History
...
- Click the Clear filters link in the Filtering card on the right
- Select the duplicate UMAP plot (with the cell colored by marker genes)
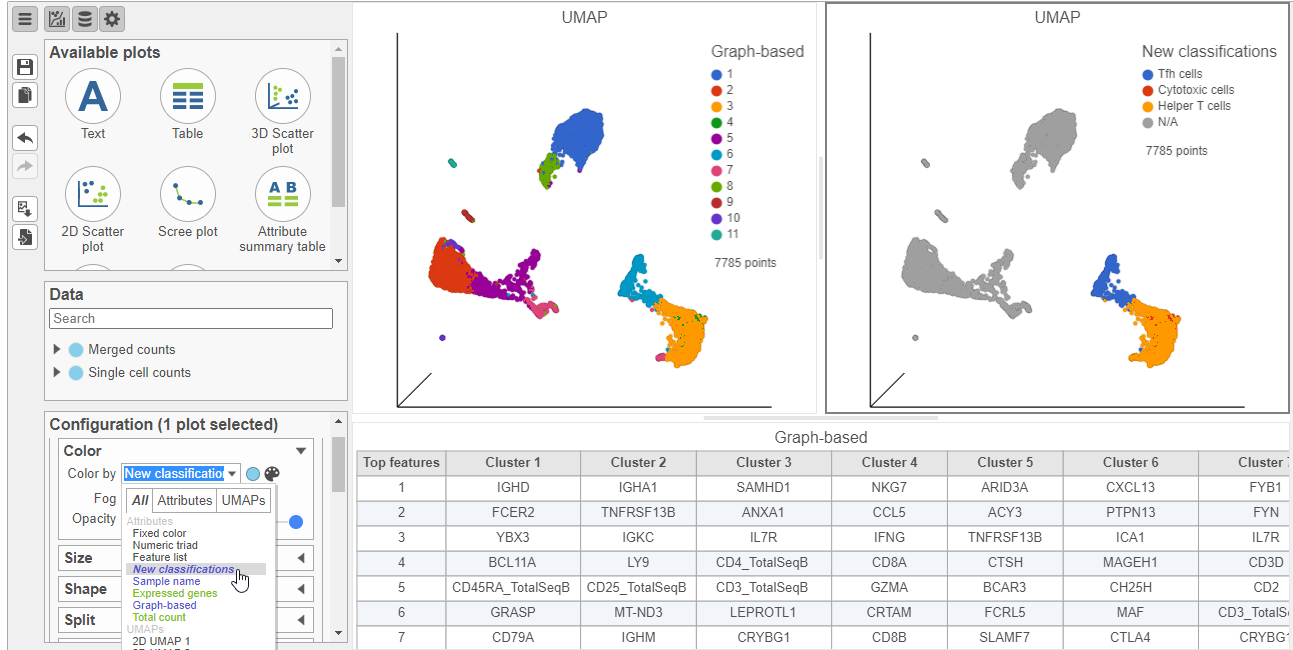
- In the Configuration card on the left, expand the Color card and color the cells by New classificationclassifications (Figure 23)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
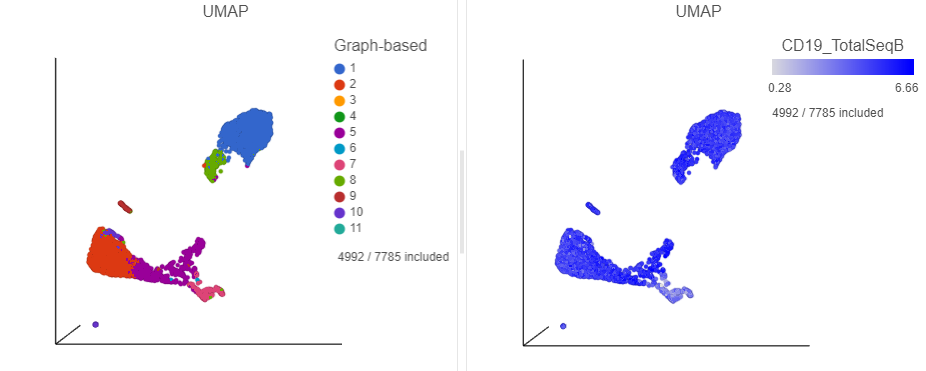
The cells in the UMAP plot are now colored from grey to blue according to their expression level for the CD19 protein marker (Figure ?24). The CD19 positive cells correspond to several graph-based clusters. We can filter to these cells to examine them more closely,
...
- Click in the top right corner of the UMAP plot
- Lasso around the CD19 positive cells (Figure ?25)
- Click in the Filtering card on the right to include the selected points
...
The plots will rescale to include the selected points. The CD19 positive cells include cells from graph-based clusters 1, 2, 5, 6, 7, 8, 9, and 10 (Figure ?26).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
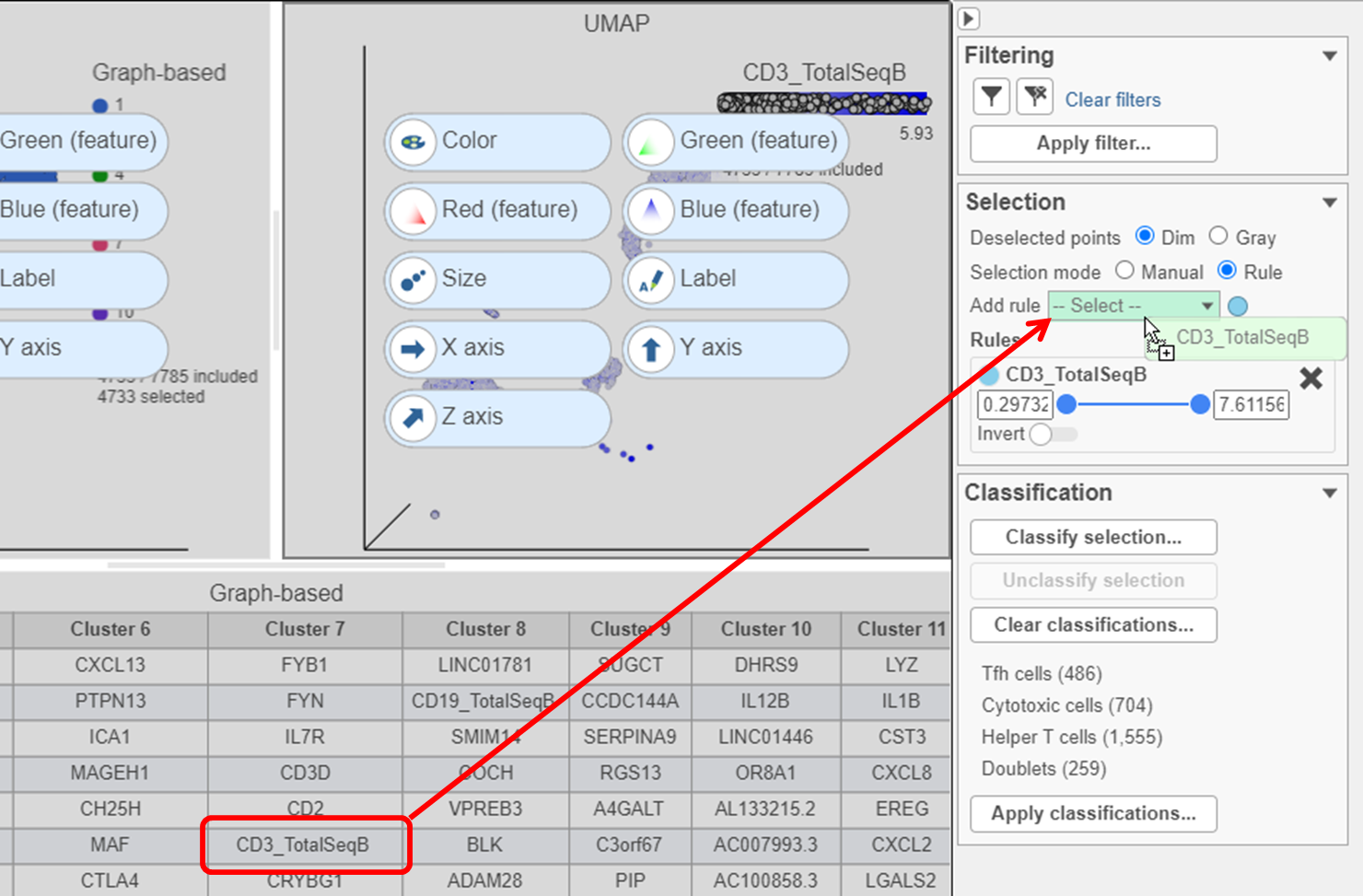
- Find the CD3_TotalSeqB protein marker in the biomarker table
- Click and drag the CD3_TotalSeqB onto the UMAP plot on the right
- Drop the CD3_TotalSeqB protein marker onto the Color configuration option on the plot (Figure 27)
While these cells express T cell markers, they also group closely with other putative B cells and express B cell markers (CD19). Therefore, these cells are likely to be doublets.
...
- Click to remove the Graph-based selection rule from the Selection card
- Find the CD3_TotalSeqB protein marker in the biomarker table
- Click and drag CD3_TotalSeqB onto the Add rule drop-down list in the Selection card (Figure ?28)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
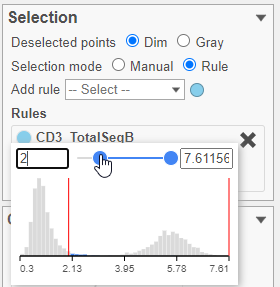
- Set the minimum threshold to 2 in the CD3_TotalSeqB selection rule (Figure ?29)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
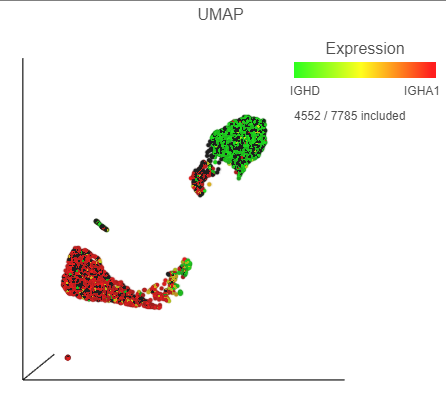
- Click, drag and drop IGHD from the biomarker table onto the Green (feature) configuration option on the UMAP plot on the right
- Click, drag and drop IGHA1 from the biomarker table onto the Red (feature) configuration option on the UMAP plot on the right (Figure 30)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
- Click in the top right corner of the UMAP plot
- Lasso around the IGHD positive cells (Figure ?31)
- In the Classification card on the right, click Classify selection
- Label the cells as Mature B cells
- Click Save
...
- Lasso around the IGHA1 positive cells (Figure ?32)
- In the Classification card on the right, click Classify selection
- Label the cells as Activated B cells
- Click Save
...
- Click the Clear filters link in the Filtering card on the right
- Select the duplicate UMAP plot (with the cell colored by marker genes)
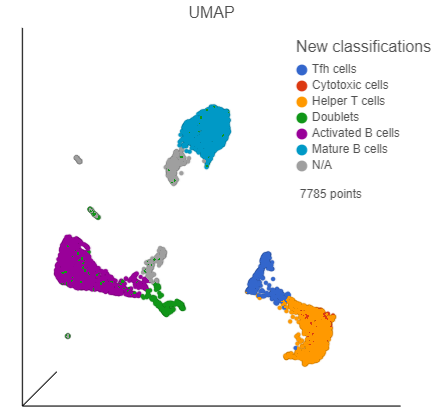
- In the Configuration card on the left, expand the Color card and color the cells by New classificationclassifications (Figure 33)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Click Apply classifications in the Classification card on the right
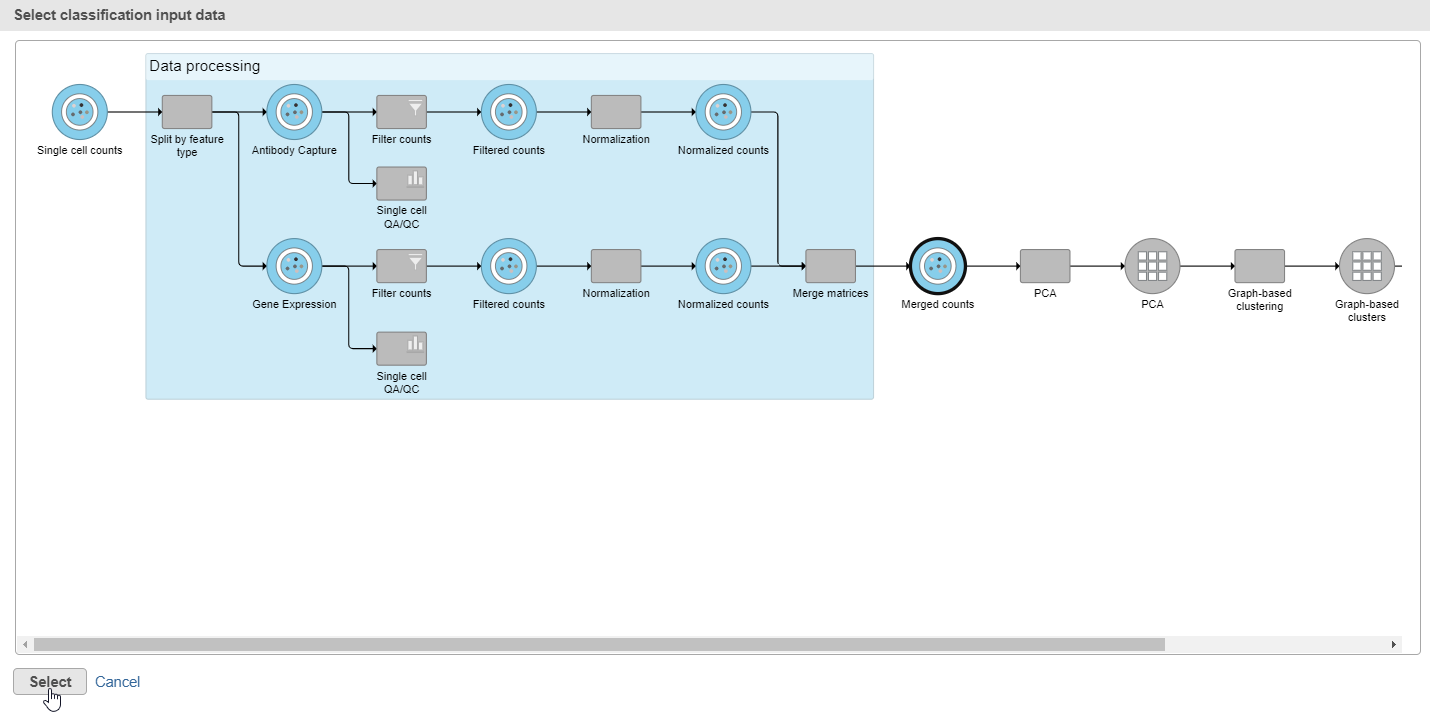
- Choose the Merged counts data node as input for the classification task (Figure ?35)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
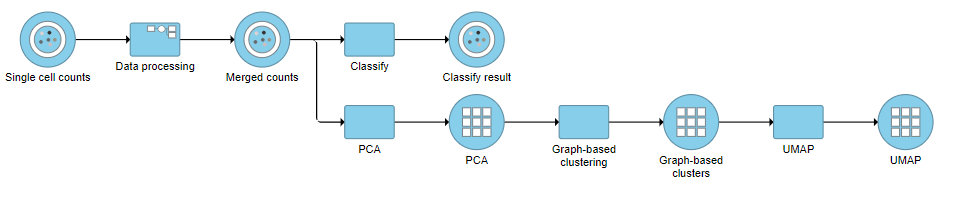
- Click the project name at the top to go back to the Analyses tab (Figure ?36)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
Overview
Content Tools