Page History
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
Filter Groups
Because we have classified our cells, we can now filter based on those classifications. This can be used to focus on a single cell type for re-clustering and sub-classification or to exclude cells that are not of interest for downstream analysis.
- Click the Classified result data node
- Click Filtering
- Click Filter groups
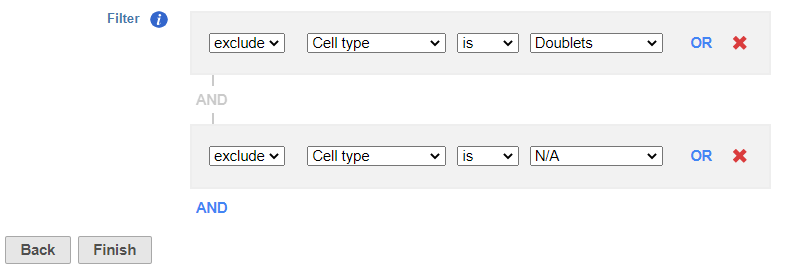
- Set to exclude
...
- Cell type is Doublets using the drop-down menus
- Click AND
- Set the second filter to exclude
...
- Cell type is N/A using the drop-down menus
- Click Finish to apply the filter (Figure ?)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
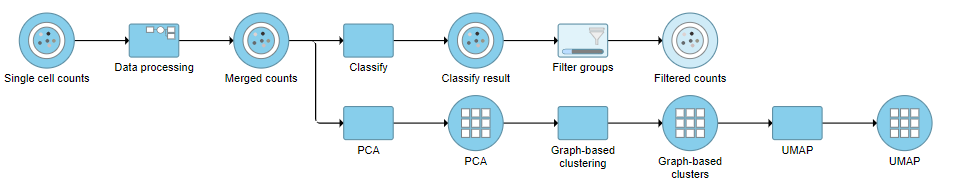
This produces a Filtered counts data node (Figure ?).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Re-split the Matrix
For this tutorial, we will re-split the protein and gene expression data prior to performing differential analysis, you may want to separate your protein and gene expression data. The split data nodes will both retain cluster and classification information.
- Click the Classified groups data node
- Click Pre-analysis tools
- Click Split matrix
This will produce two data nodes, one for each data type
Differential Analysis and Visualization - Protein Data
...
Overview
Content Tools