| Table of Contents |
|---|
| maxLevel | 2 |
|---|
| minLevel | 2 |
|---|
| exclude | Additional Assistance |
|---|
|
Section Heading
Section headings should use level 2 heading, while the content of the section should use paragraph (which is the default). You can choose the style in the first dropdown in toolbar.
We will now examine the results of our exploratory analysis and use a combination of techniques to classify different subsets of T and B cells in the MALT sample.
Exploratory Analysis Results
- Double click the UMAP data node
- In the Configuration card on the left, expand the Color card and color the cells by the Graph-based attribute (Figure ?)
| Numbered figure captions |
|---|
| SubtitleText | Color the cells in the UMAP plot by their graph-based cluster assignment |
|---|
| AnchorName | UMAP of CITE-Seq data |
|---|
|
 Image Added Image Added
|
The 3D UMAP plot opens in a new data viewer session (Figure ?). Each point is a different cell and they are clustered together in the 3D plot based on how similar their expression profiles are across proteins and genes. Because a graph-based clustering task was performed upstream, a biomarker table is also displayed under the plot. This table lists the proteins and genes that are most highly expressed in each graph-based cluster. The graph-based clustering found 11 clusters, so there are 11 columns in the biomarker table.
- Click and drag the 2D scatter plot icon from the Available plots card onto the canvas (Figure ?)
- Drop the 2D scatter plot to the right of the UMAP plot
| Numbered figure captions |
|---|
| SubtitleText | Add a 2D scatter plot and place it to the right of the UMAP plot |
|---|
| AnchorName | Add 2D scatter plot |
|---|
|
 Image Added Image Added
|
- Click Merged counts to use as data for the 2D scatter plot (Figure ?)
| Numbered figure captions |
|---|
| SubtitleText | Choose Merged counts data to draw the 2D scatter plot |
|---|
| AnchorName | Merged counts data for 2D scatter plot |
|---|
|
 Image Added Image Added
|
A 2D scatter plot has been added to the right of the UMAP plot. The points in the 2D scatter plot are the same cells as in the UMAP, but they are positioned along the x- and y-axes according to their expression level for two protein markers: CD3_TotalSeqB and CD4_TotalSeqB, respectively (Figure ?).
| Numbered figure captions |
|---|
| SubtitleText | The canvas now has a 2D scatter plot next to the UMAP |
|---|
| AnchorName | UMAP and 2D scatter plot |
|---|
|
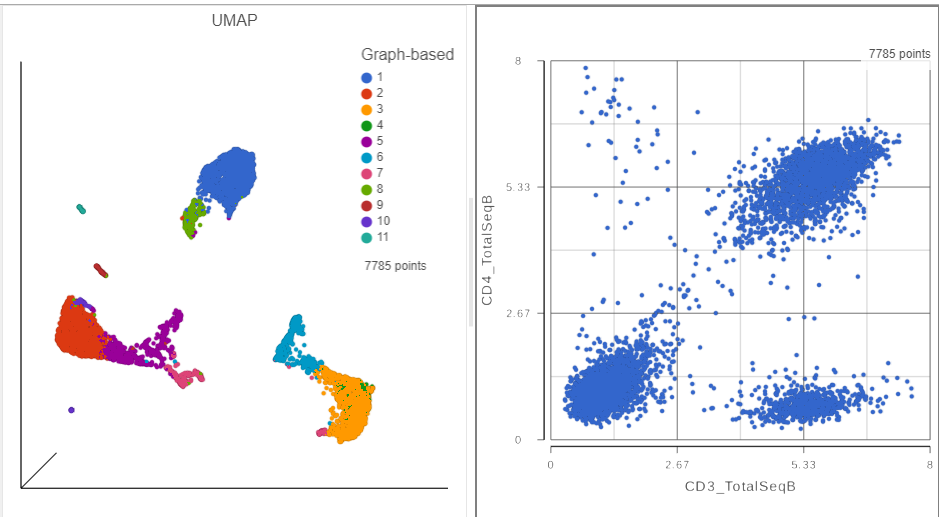 Image Added Image Added
|
- In the Selection card on the right, click Rule to change the selection mode
- Click the blue circle next to the Add rule drop-down menu (Figure ?)
| Numbered figure captions |
|---|
| SubtitleText | Click the blue circle to change the data source for the rule selector |
|---|
| AnchorName | Selection card rule mode |
|---|
|
 Image Added Image Added
|
- Click Merged counts to change the data source for the drop-down list
- Choose CD3_TotalSeqB from the drop-down list (Figure ?)
| Numbered figure captions |
|---|
| SubtitleText | Choose the CD3_TotalSeqB protein marker as a selection rule |
|---|
| AnchorName | Choose CD3 Protein marker |
|---|
|
 Image Added Image Added
|
- Click and drag the slider on the CD3D_TotalSeqB selection rule to include the CD3 positive cells (Figure ?)
| Numbered figure captions |
|---|
| SubtitleText | Use the slider to select cells with positive expression for the CD3 protein marker |
|---|
| AnchorName | Select CD3+ cells |
|---|
|
 Image Added Image Added
|
As you move the slider up and down, the corresponding points on both plots will dynamically update. The cells with a high expression for the CD3 protein marker (a marker for T cells) are highlighted and the deselected points are dimmed (Figure ?).
| Numbered figure captions |
|---|
| SubtitleText | CD3+ cells are selected on both plots |
|---|
| AnchorName | CD3+ cells selected |
|---|
|
 Image Added Image Added
|
- Click Merged counts in the Data card on the left
- Click and drag CD8a_TotalSeqB onto the 2D scatter plot (Figure ?)
- Drop CD8_TotalSeqB onto the x-axis option
| Numbered figure captions |
|---|
| SubtitleText | CHange the feature plotted on the x-axis to CD8_TotalSeqB |
|---|
| AnchorName | CD8 protein on x-axis |
|---|
|
 Image Added Image Added
|
The CD3 positive cells are still selected, but now you can see how they separate into CD4 and CD8 positive populations (Figure ?).
| Numbered figure captions |
|---|
| SubtitleText | 2D scatter plot with CD4_TotalSeqB and CD8_TotalSeqB features on the axes |
|---|
| AnchorName | CD8 and CD4 2D scatter plot |
|---|
|
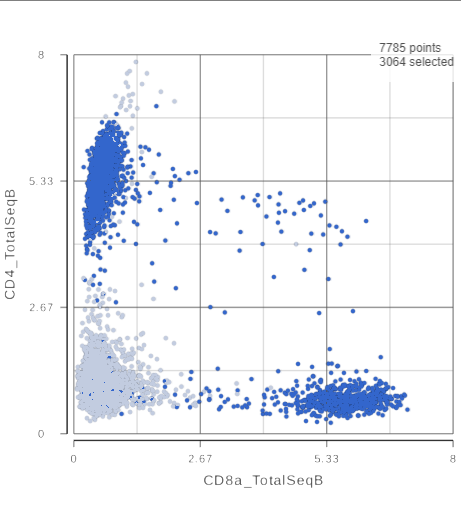 Image Added Image Added
|
Let's compare the resolution power of the corresponding CD4 and CD8A gene expression markers.
- Click the duplicate plot icon above the 2D scatter plot (Figure ?)
| Numbered figure captions |
|---|
| SubtitleText | Click the duplicate plot icon to make a copy of the 2D scatter plot |
|---|
| AnchorName | Duplicate plot |
|---|
|
 Image Added Image Added
|
- Click Merged counts in the Data card on the left
- Search for the CD4 gene
- Click and drag CD4 onto the duplicated 2D scatter plot
- Drop the CD4 gene onto the y-axis option
- Search for the CD8A gene
- Click and drag CD8A onto the duplicated 2D scatter plot
- Drop the CD8A gene onto the x-axis option
The second 2D scatter plot has the CD8A and CD4 mRNA markers on the x- and y-axis, respectively (Figure ?).
T cells
B cells
...










