Join us for an event September 26!
How to Streamline RNA-Seq analysis and increase productivity—point, click, and done
Page History
...
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
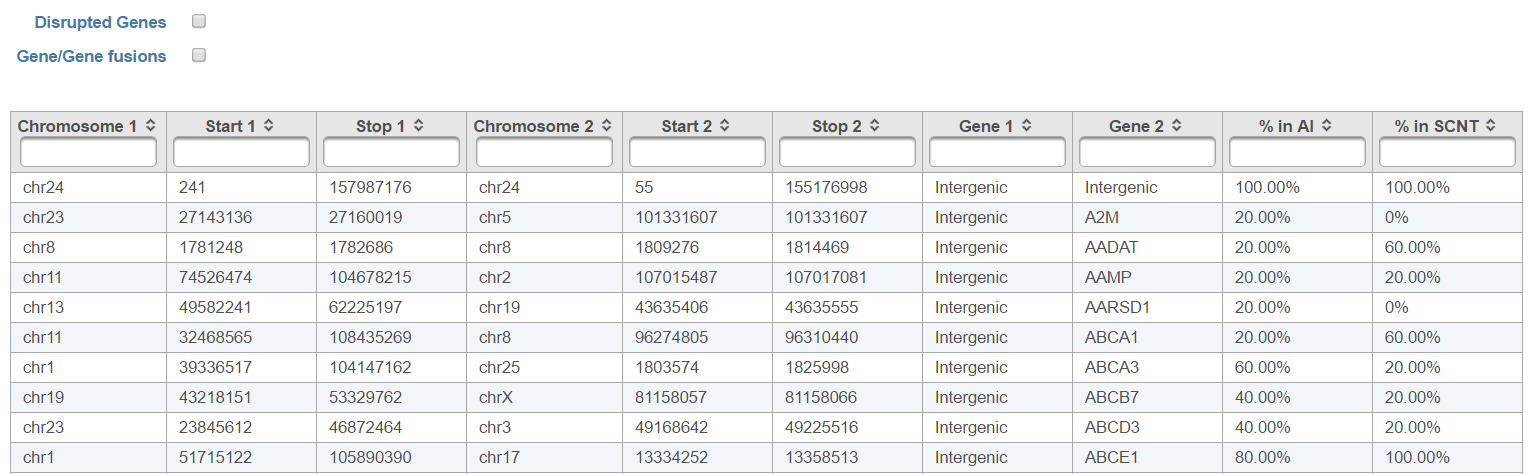
TopHat-Fusion Algorithm
General Overview
TopHat-Fusion is a version of TopHat with the ability to align reads across fusion points and detect fusion genes resulting from breakage and re-joining of two different chromosomes or from rearrangements within a chromosome (3). It is independent of gene annotation and can discover fusion products from known genes, unannotated splice variants of known genes or completely unknown genes.
...
The most up to date TopHat-Fusion version implemented in Partek® Flow® when the manual was written (2.1.0) focuses on fusions due to chromosomal rearrangements, while fusions resulting from read-through transcription or trans-splicing were not supported. For details as well as discussion of TopHat-Fusion options, see TopHat-Fusion home page (4).
Running TopHat-Fusion within Partek Flow
TopHat-Fusion is integrated in the TopHat 2 task and is invoked by using the Fusion search check box in the Alignment options dialog (Figure 1).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
STAR Algorithm
General Overview
STAR aligner also has the ability to detect fusion genes (referred to as “chimeric alignments”) (5). During the first phase of alignment, STAR searches for maximal mappable prefixes (seeds) of sequencing reads. In the second phase, all the seeds that align within user-defined genomic windows are stitched together. If an alignment within one genomic window does not cover the entire read sequence, STAR will try to find two or more windows that cover the entire read. This essentially results in detection of fusion events, with different parts of reads aligning to distal genomic locations, or different chromosomes, or different strands.
Running STAR Chimeric Alignment within Partek Flow
STAR fusion detection algorithm is integrated with STAR aligner and fusion detection is activated by tick-marking Chimeric alignment option in the Advanced options of the aligner (the Advanced options dialog is reached via Configure link in the setup dialog). As soon as the Chimeric alignment is selected, additional options, specific to the fusion search algorithm, are shown (Figure 11). For discussion on the options details, see STAR documentation.
...