Page History
...
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
TopHat-Fusion Algorithm
General Overview
TopHat-Fusion is a version of TopHat with the ability to align reads across fusion points and detect fusion genes resulting from breakage and re-joining of two different chromosomes or from rearrangements within a chromosome (3). It is independent of gene annotation and can discover fusion products from known genes, unannotated splice variants of known genes or completely unknown genes.
...
The most up to date TopHat-Fusion version implemented in Partek® Flow® when the manual was written (2.1.0) focuses on fusions due to chromosomal rearrangements, while fusions resulting from read-through transcription or trans-splicing were not supported. For details as well as discussion of TopHat-Fusion options, see TopHat-Fusion home page (4).
Running TopHat-Fusion within Partek Flow
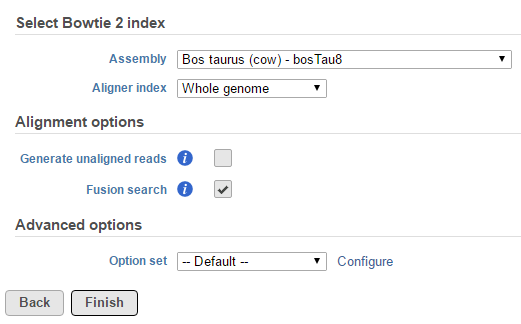
TopHat-Fusion is integrated in the TopHat 2 task and is invoked by using the Fusion search check box in the Alignment options dialog (Figure 1).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
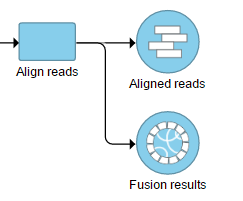
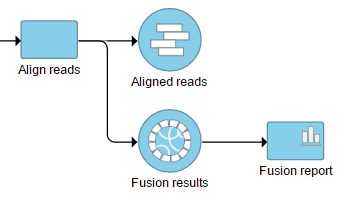
The output is generated as a new data node Fusion results (Figure 2) stemming as part of the if the TopHat 2 align reads task (in addition to Aligned reads node and, optionally, Unaligned reads node).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
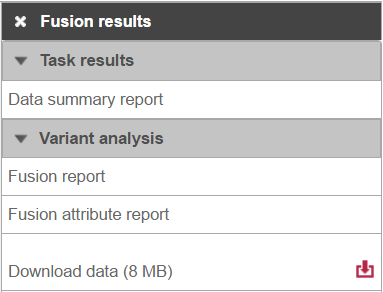
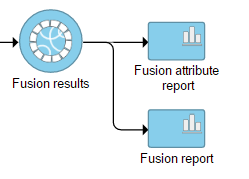
Selecting the Fusion results data node opens the task menu, with four options (Figure 3): Data summary report, Fusion report, Fusion attribute report, and Download data.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
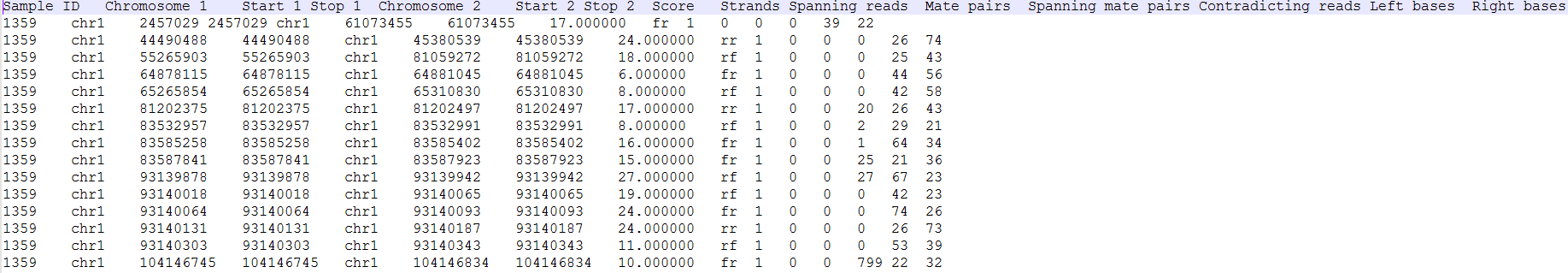
Clicking the Download data downloads a *.fusion file to the local computer. The file is human readable and can be opened in a text editor (example in Figure 4). For details refer to TopHat-Fusion documentation.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
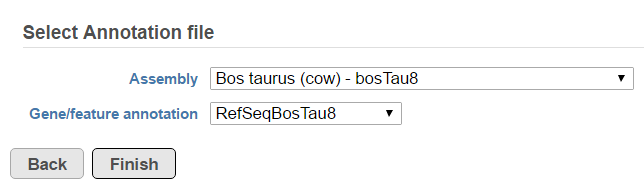
A list of annotated fusion genes, in a form of Fusion report can be obtained by first selecting the Fusion report task node (Figure 2) and then the Task report link from the task menu (Figure 3). Since the task provides an annotated report, an annotation file needs to be specified first (Figure 5).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
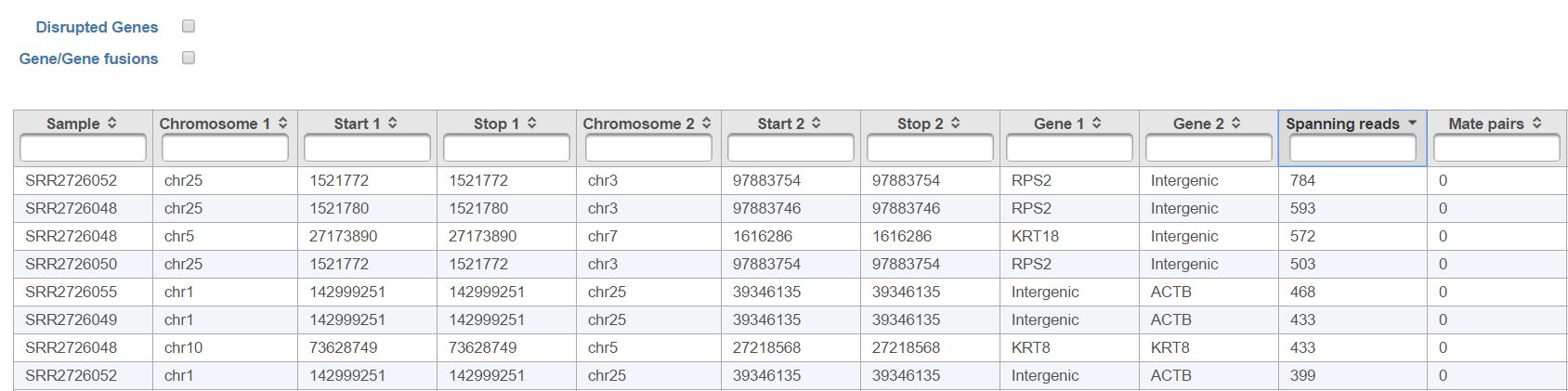
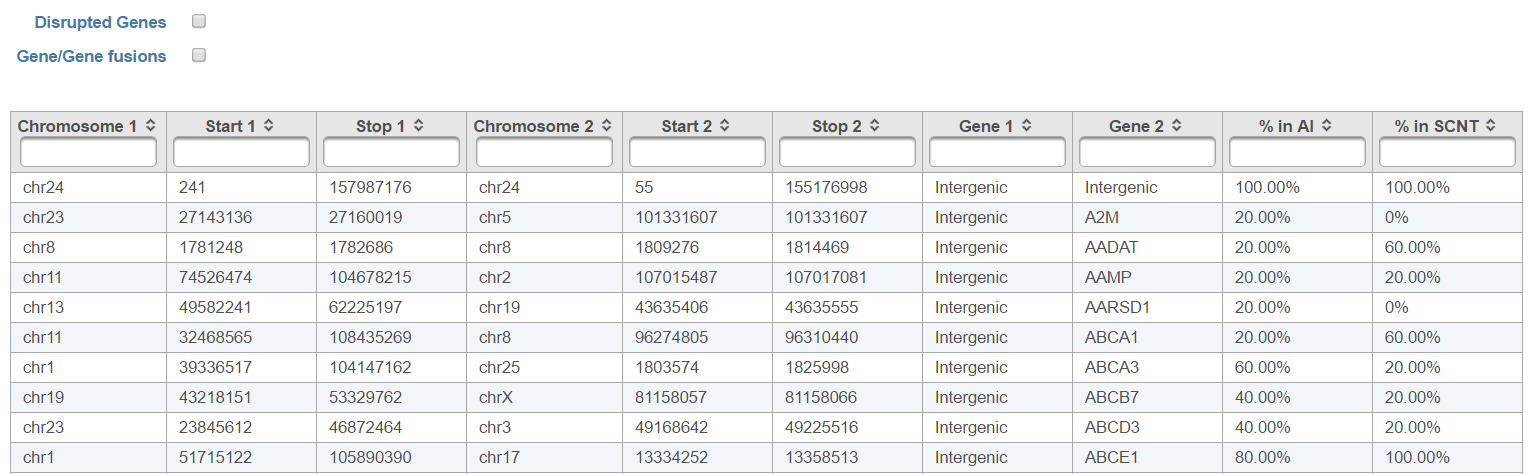
The resulting Fusion report task node (Figure 6) can be double clicked to reveal the full table (Figure 7).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
All the columns can be sorted by using the arrow buttons in column headers, while the type-in boxes can be used for searching. TopHat-Fusion does not report exact start and stop position for each side of the fusion event. It has a single location for the end of the upstream segment (Stop 1) and the beginning of the downstream segment (Start 2). Therefore, columns Start 1 and Stop 2 are added for (internal) consistency with other Partek Flow tools.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
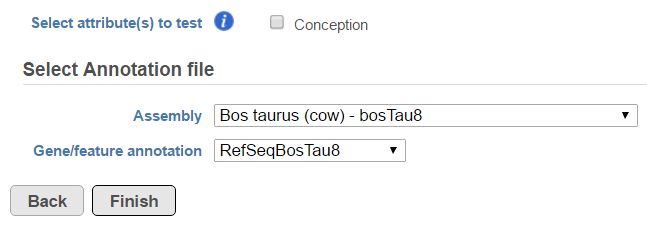
Another table which can be generated based on a Fusion results node is the Fusion attribute report (Figure 3). When the option is selected, it brings up the dialog shown in Figure 8. First, you need to specify one or more categorical attributes (Select attribute(s) to test), which have at least two categories (see Data tab). Second, you need to specify an annotation file, using the Assembly and Gene/feature annotation drop-down lists.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
A new data node, Fusion attribute report, is generated in the Analysis tab (Figure 9) and it provides access to the Task report link in the task menu.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
The checkboxes Disrupted Genes and Gene/Gene fusions are filter tools. When selected, Disrupted Genes removes all the rows (fusion events) which have no genes assigned to it, i.e. those that merge two intergenic regions. However, if there is a fusion between a gene and an intergenic region, it will be kept in the table. The Gene/Gene fusions filters in only those fusion events which have an annotated gene on both sides of the breakpoint. In the other words, only gene to gene fusions are kept in the table.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
STAR Algorithm
General Overview
STAR aligner also has the ability to detect fusion genes (referred to as “chimeric alignments”) (5). During the first phase of alignment, STAR searches for maximal mappable prefixes (seeds) of sequencing reads. In the second phase, all the seeds that align within user-defined genomic windows are stitched together. If an alignment within one genomic window does not cover the entire read sequence, STAR will try to find two or more windows that cover the entire read. This essentially results in detection of fusion events, with different parts of reads aligning to distal genomic locations, or different chromosomes, or different strands.
Running STAR Chimeric Alignment within Partek Flow
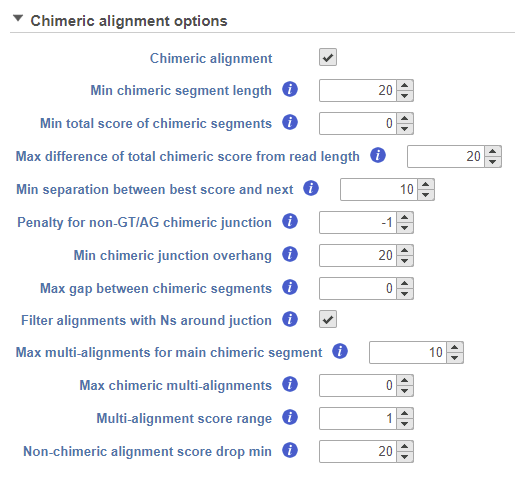
STAR fusion detection algorithm is integrated with STAR aligner and fusion detection is activated by tick-marking Chimeric alignment option in the Advanced options of the aligner (the Advanced options dialog is reached via Configure link in the setup dialog). As soon as the Chimeric alignment is selected, additional options, specific to the fusion search algorithm, are shown (Figure 11). For discussion on the options details, see STAR documentation.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
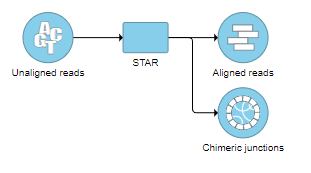
The output is associated with the Chimeric results data node (Figure 12), which is a part of STAR results (in addition to Aligned reads node and, optionally, Unaligned reads node).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Selecting the Chimeric results node opens the task menu (Figure 13) with twooptions: Data summary report or Download data.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
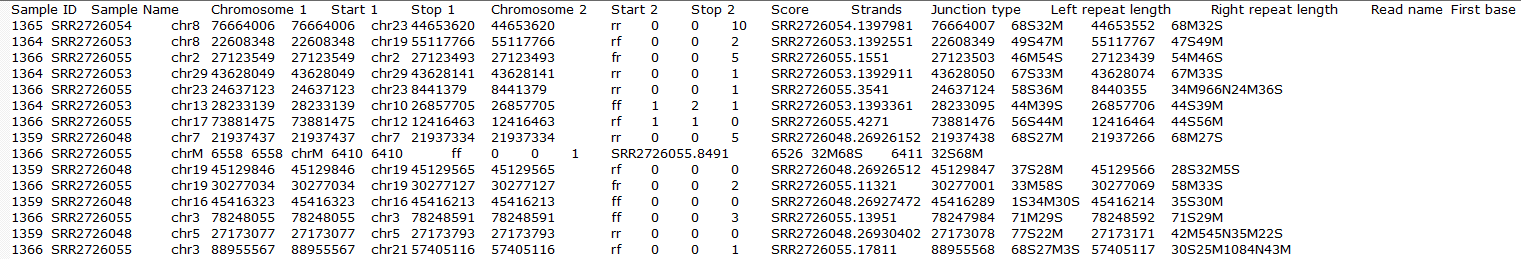
Clicking on the Download data results in download of a .fusion file to the local computer. The file is human readable and can be opened in a text editor (example in Figure 14). For details refer to STAR's documentation.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
References
- Annala MJ, Parker BC, Zhang W, Nykter M. Fusion genes and their discovery using high throughput sequencing. Cancer Lett. 2013;340:192-200.
- Costa V, Aprile M, Esposito R, Ciccodicola A. RNA-Seq and human complex diseases: recent accomplishments and future perspectives. Eur J Hum Genet. 2013;21:134-142.
- Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biology. 2011;12:R72
- TopHat-Fusion. An algorithm for discovery of novel fusion transcripts. http:// http://tophat.cbcb.umd.edu/fusion_index.html Accessed on April 25, 2014
- Dobin A, Davies CA, Schlesinger F et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15-21.
...
...
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|
...