Page History
...
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
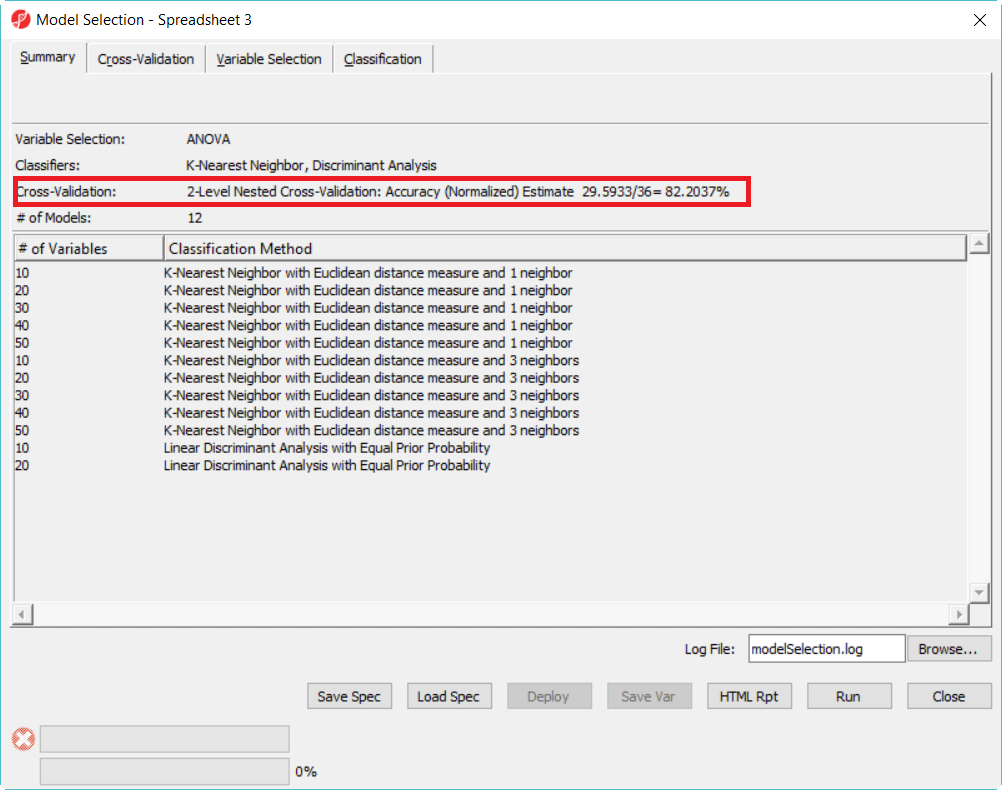
Select a Classification Model
Download the zip file from the link below. The download contains the following files:
...
- Click on Deploy button to leaunch the model using the whole dataset, but first save the file as 20var-1NN-Euclidean.ppb. It will run ANOVA on the 28 samples to generate the top 20 genes and build a model using 3 K-Nearest neighbor based on Euclidean distance measure.
- Since the deployed model was from the whole 28 samples, in order to know the correct rate, we need a test set to run the model on.
Deploying a Model
To get unbiased correct rate, the test set sample must be independent from the training set. Now we are going to load another dataset, it has 8 samples with logged intensity values on the set of genes as that of the training data set. To use a complete independent test set to get correct rate is called hold-out validation.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
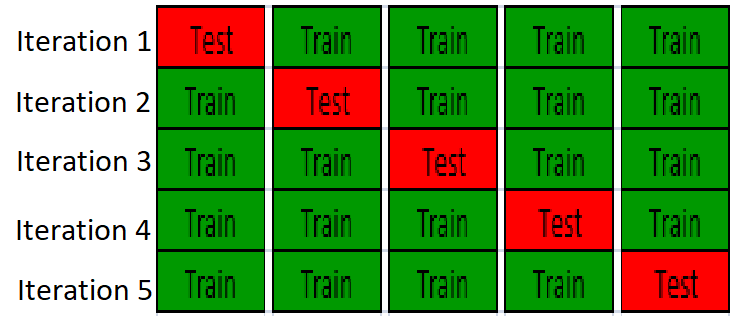
Cross-validation
Cross validation is used to esimate the accuracy of the predictive model, it is the solution to overfitting problem. One example of ovefitting is testing the model on the same data set when the model is trained. It is like to give sample questions to students to practice before exam, and the exact same questions are given to the students during exam, so the result is biased.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Common Mistakes
In Partek model selection tool, the cross-validation is performed first. Each iteration of cross-valiation, the variable selection and classification are performed on the training set, and the test set is completely independent to validate the model. One common mistake is to select variable beforehand, e.g. using perform ANOVA on the whole dataset and use ANOVA's result to select top genes, and perform the cross-valiation to get correct rate. In this case, the test sets in cross validation were used in the variable selection, it is not independend from the training set, so the result will be biased.
...