Page History
...
Download the zip file from the link below. The download contains the following files:
- training Training set data–28 samples (11 disease samples and 15 normal samples) on 9953 genes
- test Test set data – 8 samples on 9953 genes
- configuration of the model builder (.pcms file)
- deployed model (.pbb file)
...
- Open Partek Genomics Suite, choose File>Open.. from the main menu to open the trainingSet Training.fmt.
- Select Tools > Predict > Model Selection from the Partek main menu
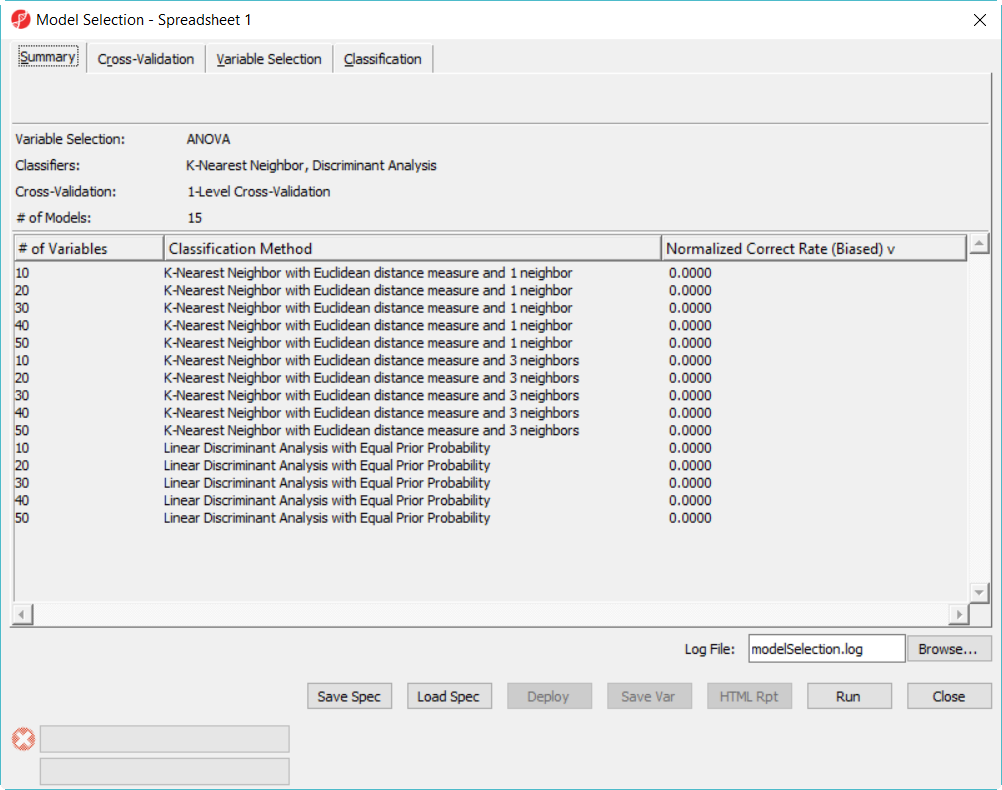
- In Cross-Validation tab, choose to Predict onType, Positive Outcome is Disease, Selection Criterion is Normalized Correct Rate (Figure 1)
- Choose 1-Level Cross-Validation option, and use Manually specify partition option as 5– use 1-level cross validation option is to select the best model to deploy
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
When click on Run, a dialog as (Figure 4) will display, some classifiers like discriminant analysis are not recommended to perform on dataset with more number of variables than that of samples.
...
- Click on Deploy button to deploy the model using the whole dataset, save the file as 20var-3NN1NN-Euclidean.ppb. It will run ANOVA on the 28 samples to generate the top 20 genes and build a model using 3 K-Nearest neighbor based on Euclidean distance measure.
- Since the deployed model was from the whole 28 samples, in order to know the correct rate, we need a test set to run the model on.
...
Another method to get unbiased accuracy estimate in Partek Genomics Suite is to do 2-Level Cross validation. You can use the whole The following steps is to show how to use all the 36 samples to select the best model and the accuracy estimate using all the 36 samples. 36 sample data set --combining both training set and test set samples, in this example is to use the 36 samples.
- Choose File>Open... to open to browse and open 36samples.fmt
- Choose Tools>Predict>Model Selection... from the menu
- Click on Load Spec to select tutorial.pcms
- Click Run on 1-level cross validation to select the best model using 36 samples
The best model is 30 variables using 1-Nearest Neighbor with Euclidean distance measure
Cross validation
Common mistakes
...