...
- Under the Visualization section in the Gene Expression workflow, select Cluster Based on Significant Genes(Figure 1)
| Numbered figure captions |
|---|
| SubtitleText | Select cluster based on significant genes from the Visualization panel of the Gene Expression workflow |
|---|
| AnchorName | Selecting Clustering |
|---|
|
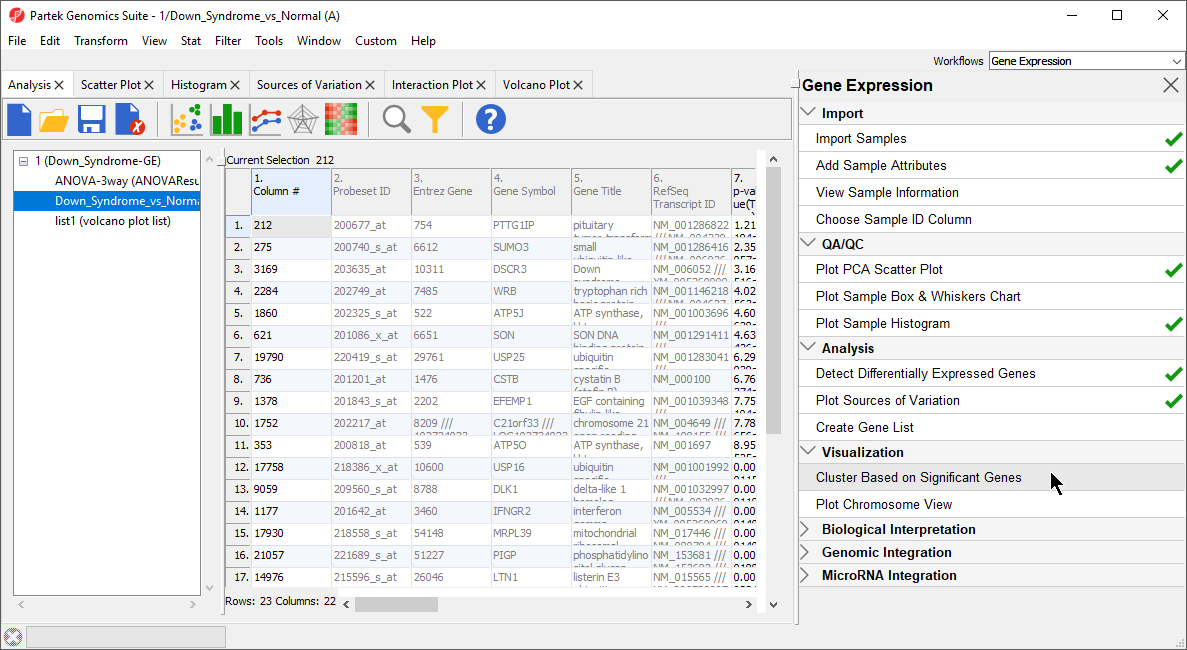 Image Removed Image Removed
|
- The Cluster Significant Genes dialog asks you to specify the type of clustering you want to perform. Select Hierarchical Clustering and select OK
- Choose the Down_Syndrome_vs_Normal (A) spreadsheet under the Spreadsheet with differentially expressed genes
- Choose the Standardize – shift genes to mean of zero and scale to standard deviation of one under the Expression normalization panel . (Figure 1)
This option will adjust all the gene intensities such that the mean is zero and the standard deviation is 1
| Numbered figure captions |
|---|
| SubtitleText | Configuring Hierarchical Clustering |
|---|
| AnchorName | Hierarchical Clustering options |
|---|
|
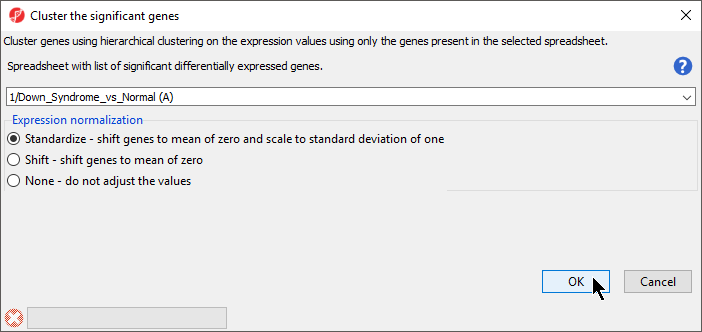 Image Added Image Added
|
- Select OK to generate a Hierarchical Clustering tab (Figure 2)
| Numbered figure captions |
|---|
| SubtitleText | Hierarchical Clustering of Down_Syndrome_vs_Normal (A) |
|---|
| AnchorName | Hierarchical Clustering |
|---|
|
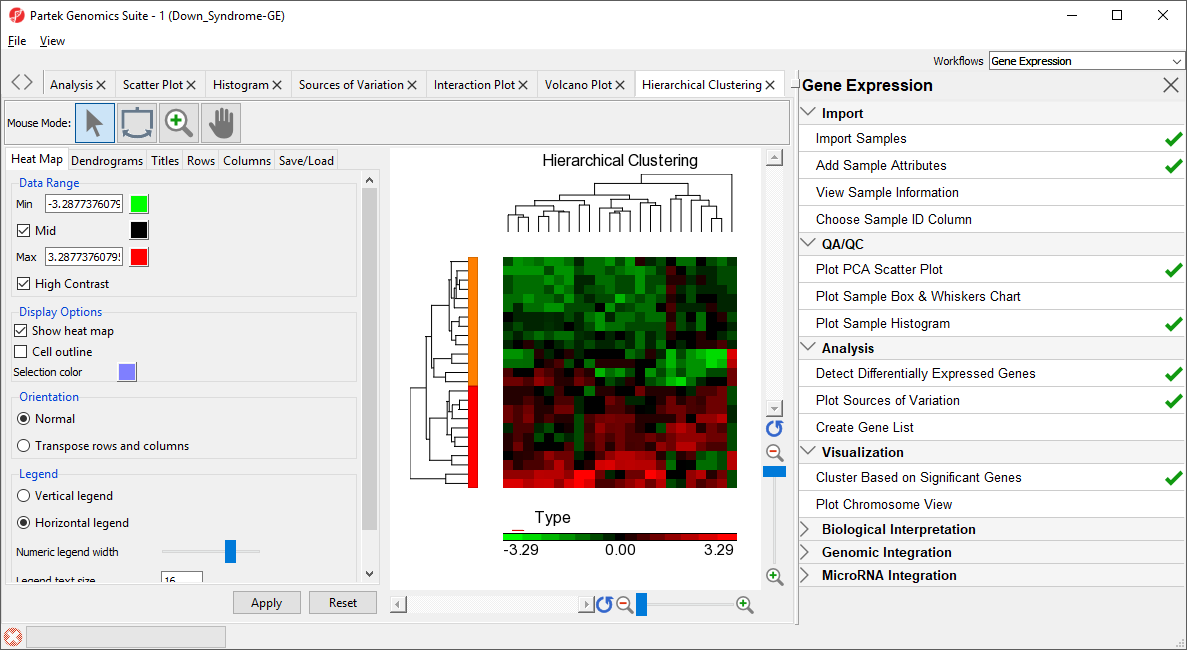 Image Removed Image Removed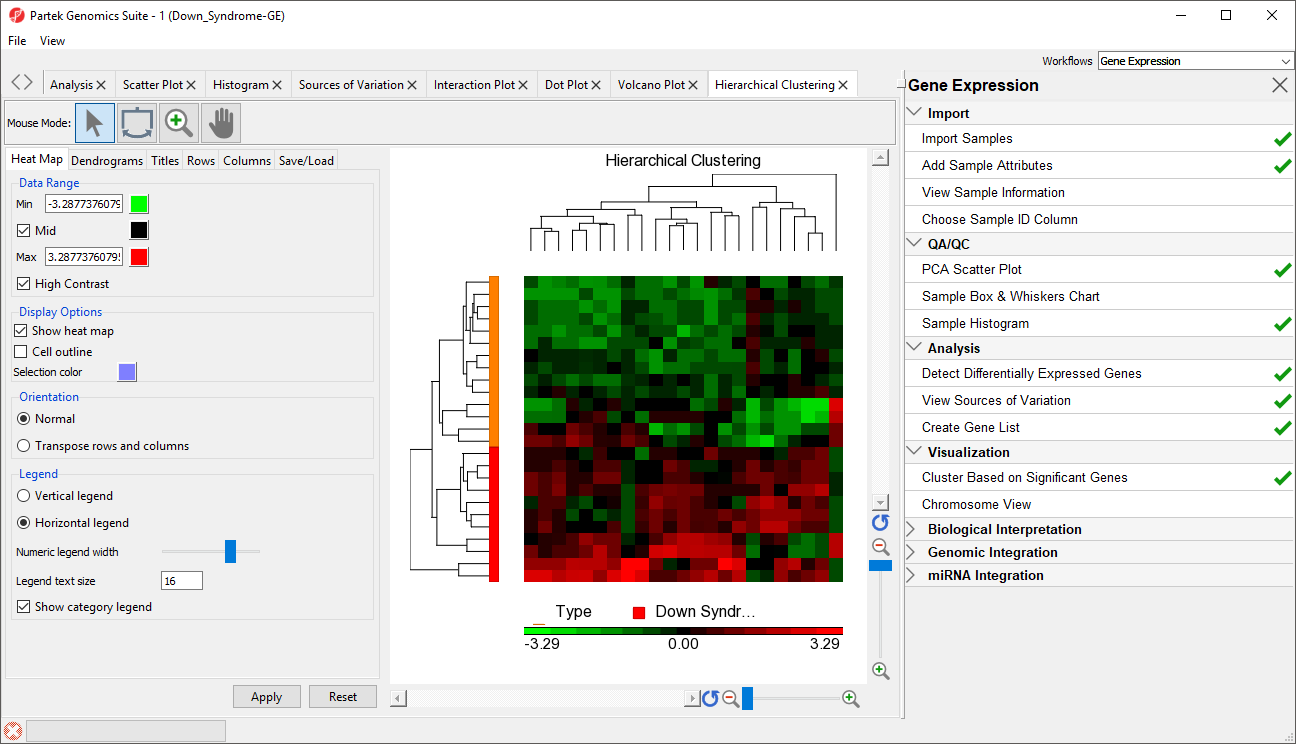 Image Added Image Added
|
The graph (Figure 2) illustrates the standardized gene expression level of each gene in each sample. Each gene is represented in one column, and each sample is represented in one row. Genes with no difference in expression have a value of zero and are colored black. Genes with increased expression in Down syndrome samples have positive values and are colored red. Genes with reduced expression in Down syndrome samples have negative values and are colored green. Down syndrome samples are colored red and normal samples are colored orange. On the left-hand side of the graph, we can see that the Down syndrome samples cluster together.
...



