Page History
Table of Contents maxLevel 2 minLevel 2 exclude Additional Assistance
Importing a region list
A region list must contain the chromosome, start location, and stop locations as the first three columns. The chromosome number in the region list must be compatible with the genomic annotation for the species if you plan to use any feature (like motif detection) that requires reference sequence information.
- Import the region list as described above for text files . with the following options
- Select Other for data type
- Set chromosome as a text field
- Set location start and stop
- as either integer or
- text fields
- Right-click on the imported spreadsheet in the spreadsheet tree
- Select Properties
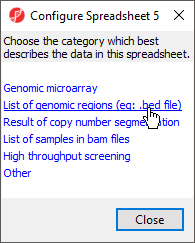
- Select List of genomic regions from the Configure Spreadsheet dialog to add region to the properties
- (Figure 1)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The spreadsheet properties will now include region. Alternatively, region can be added as a spreadsheet property from the Configure Genomic Properties dialog by selecting Advanced.. , choosing region from the drop-down menu, selecting Add, and selecting OK.
If you would like to do any operation that requires looking up hte the reference genomic sequence information for the regions based on genomic location, you will need to specify the species for this region list.
- Right-click on the imported spreadsheet in the spreadsheet tree
- Select Properties
- Select Genomic from the Add Property drop-down menu
- Select Add
- Select the source Species and Genome Build from the drop-down menus
- Select OK
With a few additional options, the region list can be made viewable in the genome browser.
- Right-click on the imported spreadsheet in the spreadsheet tree
- Select Properties
- Select Advanced..
- Select Edit next to the species name
- Specify the Cytoband file and 2Bit sequence file
- Select OK
Motif detection
Starting with a region list, you may detect either known or de novo motifs using the ChIP-Seq
Seq workflow if your spreadsheet has been associated with a species and a reference genome as
described in the import section.
- Select ChIP-Seq from the Workflows drop-down menu
- Select Motif detection from the Peak Analysis section of the workflow
Both options ( Discover de novo motifs and Search for known motifs) can now can be performed. Motifs (de novo) may be displayed in the Genome Browser and known detected motifs may be viewed in web-log format by right-clicking on a header row of the motif spreadsheet.
Determining the average values for a region list
If you have a region list or a bed .BED file and you have a microarray experiment with data, you can summarize the data according to the genomic coordinates contained in the region list. For instance, the region list contains a list of CpG islands, the experiment contains methylation percentage values for probes (β values), and you would like to summarize the methylation values for individual probes for the CpG islands. Or you have a list of copy number amplifications, microarray gene expression data, and you are interested in determining if the average intensities of the probes in those regions is higher than expected.
Import the region list (or BED file) and specify the region property as explained elsewhere in this document
- With the region list spreadsheet selected, right-click in any column header and select Insert Average
A dialog box similar to that shown. With the Add Average tab selected, specify the location where you like the new columns to appear by using the Add to the and of Column pull-down menus. Specify the top-level spreadsheet containing the data you wish to be averaged (β values, gene-intensity values, etc.) in the Get average from spreadsheet pull-down menu. Choose the radio button to specify how the averaging should be done. The bottom two choices (Mean of all samples and Mean value for all samples separately) are obvious; the first option (Mean of samples significant in region) is used when the region list has a SampleID associated with each region. In this case, the column designated as the SampleID column from the top-level spreadsheet will be used to identify the sample to be summarized for each region.
Find region overlaps
You have a list of regions from another analysis program (perhaps you detected peaks using an R program) and you’d like to compare that region list with a region list that Genomics Suite calculated. Perhaps you have two lists created by Genomics Suite (one generated from peak detection with one set of parameters and the other created with different parameters) and you’d like to see what the two lists have in common. You may use the Tools > Find Region Overlaps command to compare two or more region lists as shown.
...
In contrast, the second option, Only report regions present in all lists, will intersect both lists and only reports regions found in all the lists.
Importing genomic locations to be used with annotating SNVs
The Tools > Annotate SNVs feature requires four columns of data per genomic location: the position of the SNP (chr.basePosition), the SampleName, a reference base, and the SNP call (single nucleotide or genotype) as shown in Figure 20.
...