Page History
...
There are many useful visualizations, annotations, and biological interpretations that can operate on a gene list. In order for these features operate on an imported list, an annotation file must first be associated with the gene-list. Additionally, many operations that work with a list of significant genes (like GO- or Pathway-Enrichment) require comparison against a background of “non-significant” genes. The quickest way to accomplish both is to use the background of “all genes” for that organism provided by an annotation source like RefSeq, Ensembl, etc. in .pannot (Partek® annotation), .gff, .gtf, .bed, tab- or comma-delimited format. If the file is not already in a tab-separated or comma delimited format, you may import, modify, and save the file in the proper file format.
...
Associating a spreadsheet with an annotation file
- Select File from the main toolbar
- Select Genomic Database under Import (Figure 1)
...
The annotation file has been associated with the spreadsheet and additional tasks can now be performed on the data.
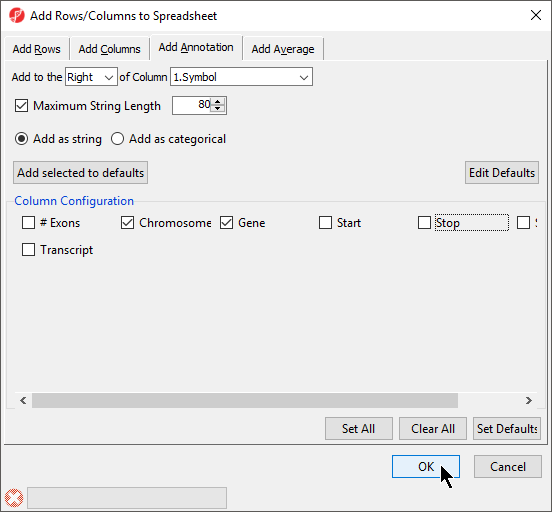
Adding annotations to a spreadsheet
Inserting annotations from an annotation file
If an annotation file has been associated with a spreadsheet, annotations from the file can be added as columns in the spreadsheet.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Annotating with cytobands
- Select Annotate with Cytobands from Tools in the main toolbar when a suitable spreadsheet is open
A column with cytoband locations will be added to the spreadsheet. Adding a cytoband is possible if genomic coordinates are associated with the gene list spreadsheet during import or by association with an annotation file.
Annotating with known SNPs
- Select Annotate with Known SNPs from Tools in the main toolbar when a suitable spreadsheet is open will add
...