...
- For this tutorial, unzip the files to C:\Partek Training Data\Down_Syndrome-GE or to a directory of your choosing. Be sure to create a directory or folder to hold the contents of the zip file
- Copy or move the annotation files (HG-U133A.cdf, HG-U133A.na36.annot, HG-U133A.na36.annot.idx) to C:\Microarray Libraries. (Copying the annotation files to the default library location is done because newer annotation files that are released after the publication of this tutorial may cause the results to be different than what is shown in the published tutorial. If, however, you prefer to download the latest version, you may omit copying the HG-U133A files to C:\Microarray Libraries)
- Start PGS and select Gene Expression from the Workflows panel on the right side of the tool bar in the PGS main window (Figure 1)
| Numbered figure captions |
|---|
| SubtitleText | Selecting the gene expression workflow |
|---|
| AnchorName | Gene Expression Workflow |
|---|
|

|
| Numbered figure captions |
|---|
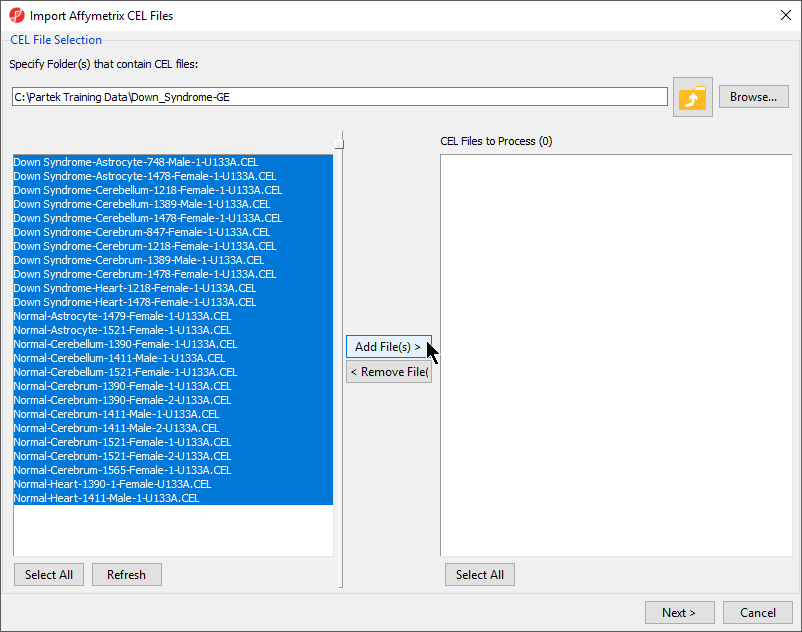
| SubtitleText | Selecting the folder and CEL files for the experiment |
|---|
| AnchorName | CEL File Selection |
|---|
|
|
- Select the Add File(s) > button to move all the .CEL files to the right panel. Twenty-five CEL files will be processed
- Select the Next > button to open the Import Affymetrix CEL Files dialog (Figure 3)
| Numbered figure captions |
|---|
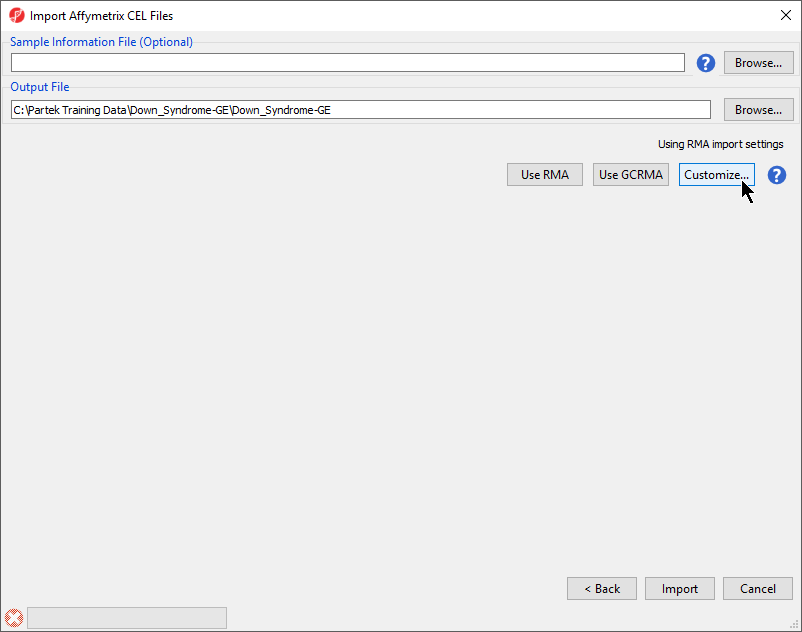
| SubtitleText | Configuring import files window |
|---|
| AnchorName | Import Affymetrix CEL Files |
|---|
|
|
| Numbered figure captions |
|---|
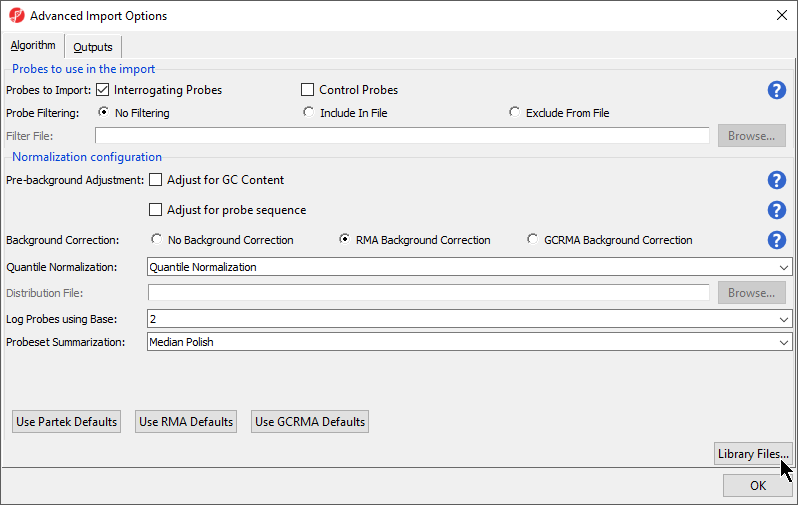
| SubtitleText | Configuring the Advanced Import Options dialog |
|---|
| AnchorName | Advanced Import Options |
|---|
|
|
- Select Library Files… to open the Specify File Locations dialog (Figure 5). This dialog is used to specify the location of the library folder and the annotation files
...
- The default library location can be modified at by selecting Change... in the Default Library File Folder panel. By default, the library directory is at C:\Microarray Libraries. This directory is used to store all the external libraries and annotation files needed for analysis and visualization. The library directory can also be modified from Tools > File Manager in the main PGS menu
- Select OK (Figure 5) to close the Specify File Locations dialog
- Select the Outputs tab from the Advanced Import Options dialog (Figure 6)
| Numbered figure captions |
|---|
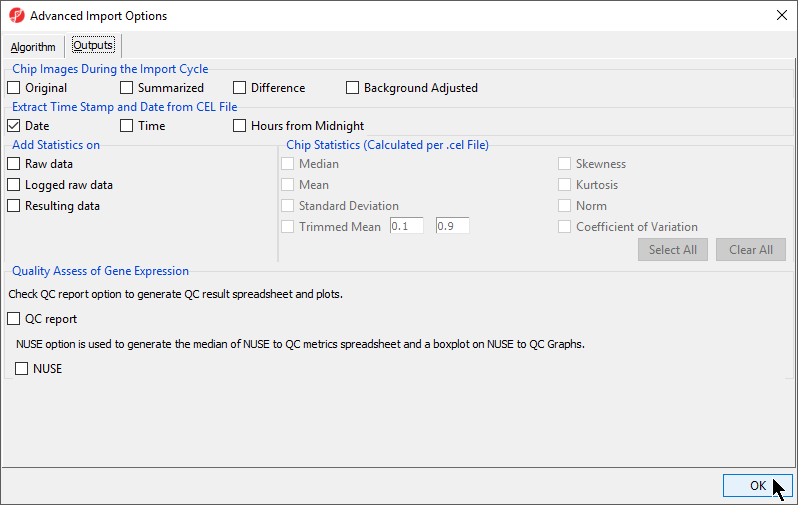
| SubtitleText | Specifying Advanced Import Options to create chip images of and extract the scan date from the CEL files |
|---|
| AnchorName | Advanced Import Options Outputs |
|---|
|
|
...
After importing the .CEL files has finished, the result file will open in PGS as a spreadsheet named 1 (Down_Syndrome-GE). The spreadsheet should contain 25 rows representing the micoarray chips (samples) and over 22,000 columns representing the probe sets (genes) (Figure 7).
| Numbered figure captions |
|---|
| SubtitleText | Viewing the main or top-level spreadsheet |
|---|
| AnchorName | Viewing Imported Data |
|---|
|

|
For additional information on importing data into PGS, see
Chapter 4 Importing and Exporting Data in the Partek User’s Manual. The User’s Manual is available from the Partek Genomic Suite menu from
Help> User’s Manual. The
FAQ (
Help > On-line Tutorials > FAQ) may also be helpful. As this tutorial only addresses some topics, you may need to consult the User’s Manual for additional information about other useful features.
...





